Research Projects
Project 1: Development of free energy calculation methods for biomolecular associations
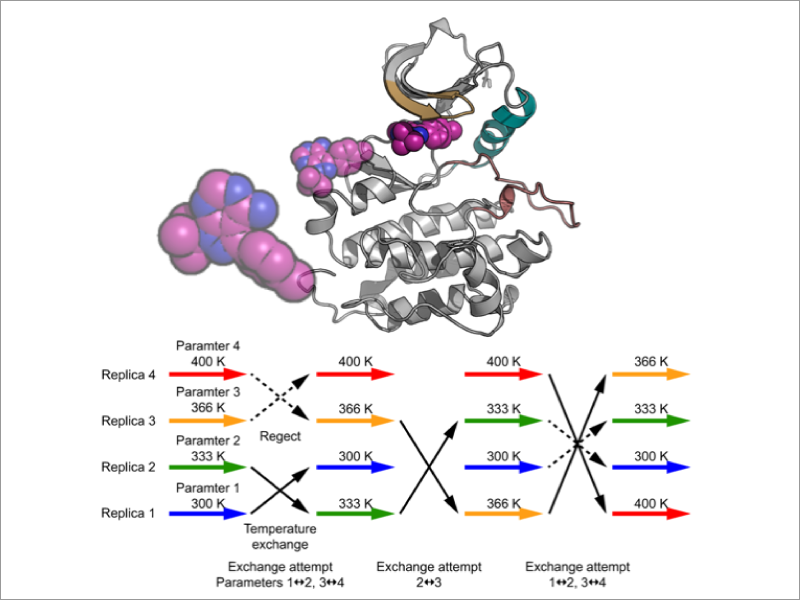
We aim to predicts free energy changes (both absolute and relative changes) accompanying protein-ligand and protein-protein association with high precision and efficiency. For this purpose, we develop computational methods based on both enhanced sampling methods (such as the Replica-Exchange MD method) and free energy perturbation methods, which can effectively utilize supercomputers and GPU parallel computers.
Project 2: Multiscale modeling of crowded cellular environments
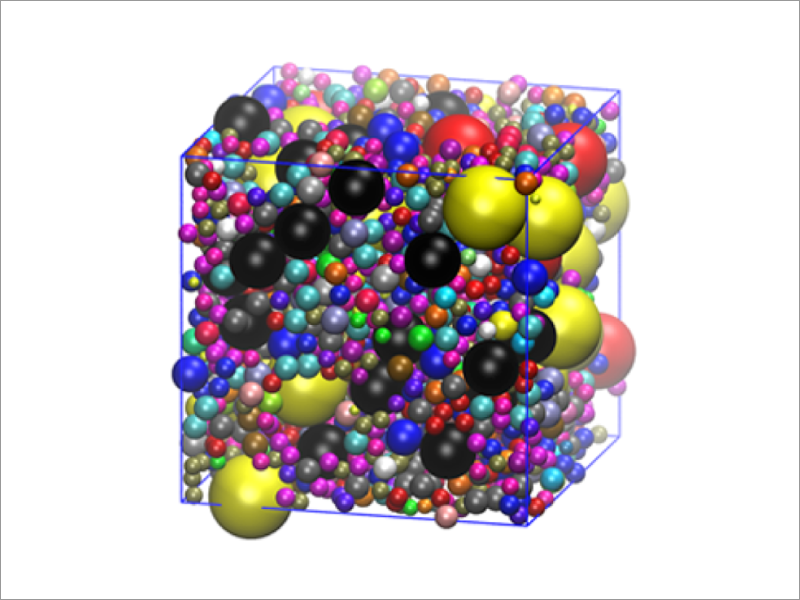
Cellular environment is significantly different from in vitro dilute environment. Inside the cell, the concentration of macromolecules, such as proteins and nucleic acids is in the range of 80–400 mg/mL. In order to carry out simulations in intracellular environment, we need to consider a system containing a large number of atoms and simulate for an extended amount of time. To speed up simulations, we develop multiscale models of proteins, nucleic acids and membranes, including an efficient coarse-grained model, and implement them in GENESIS.
Project 3: Understanding protein functions relevant for signal transduction for the purpose of drug discovery
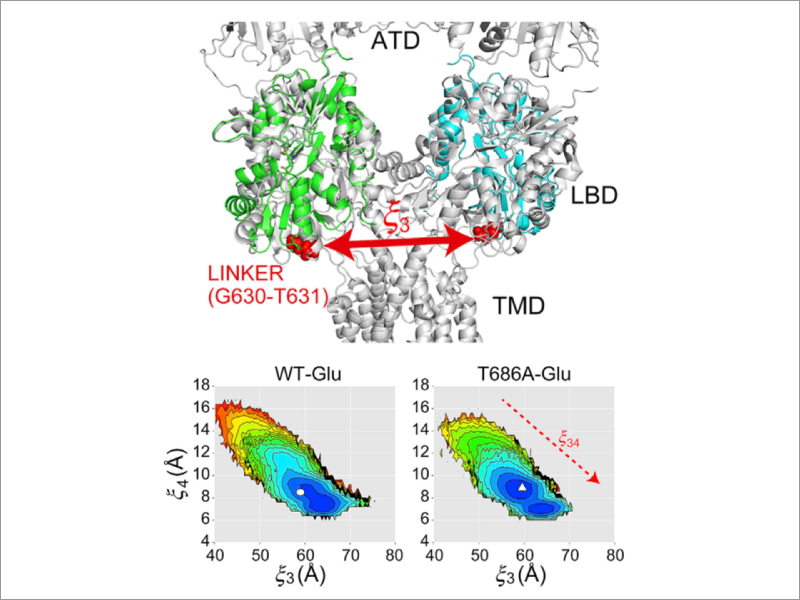
We aim to understand the molecular mechanism of diseases such as cancer mechanisms. The obtained knowledge can be applied for the purpose of drug discovery. To this end, we apply cutting-edge simulation methods to reveal atomic details of protein-ligand and protein-protein interactions regulating the various signal transduction processes. Combined with largescale MD simulations, we aim to understand molecular basis of cellular functions in collaboration with experimental groups in the fields of structural biology, molecular cellular biology, and single molecule experiments.
Gallery
Movies and figures in our research activity
Facilities
Research facilities in our laboratory