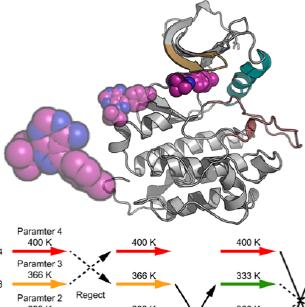
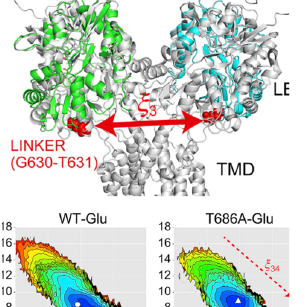
We develop computational methods to investigate, at atomic level, the structure-dynamics-function relationship of biomolecules in cells, using massively parallel molecular dynamics simulation program GENESIS. By applying the developed methods, and through collaboration with experimental groups, we aim to understand molecular recognitions underlying a various signaling process.
Events & NewsMore
Message
In this laboratory, we study biomolecular functions in a living cell, in particular, protein functions in signal transduction pathways, using theoretical/computational chemistry. For this purpose, we aim to understand and predict protein-ligand or protein-protein binding processes in dilute solution or crowded cellular environments.
Lead Researcher: Yuji Sugita (Laboratory for biomolecular function simulation)