Cells regulate their function by receiving external signals through receptors in the cell membrane, and transmitting them to the cytoplasm and nucleus. Dysregulation of signal transduction due to malfunctioning proteins is the cause of several diseases. For example, in cancer, cell proliferation signals are dysregulated by abnormal proteins involved in the signaling pathway. There is an ongoing development of molecular target drugs that inhibit ligand binding to abnormal proteins to stop their function. We reveal atomic details of protein-ligand bindings and protein-protein interactions related to signal transduction by MD simulation and free energy analyses for the purpose of drug discovery. In addition to performing extensive simulations, we also work with different experimental groups in the fields of structural biology, molecular cell biology, and single molecule measurements to understand cellular function at the molecular level by bottom-up approaches.
References
- Encounter complexes and hidden poses of kinase-inhibitor binding on the free-energy landscape. Suyong Re, Hiraku Oshima, Kento Kasahara, Motoshi Kamiya, and Yuji Sugita. Proc. Natl. Acad. Sci. USA, 116, 18404-18409 (2019).
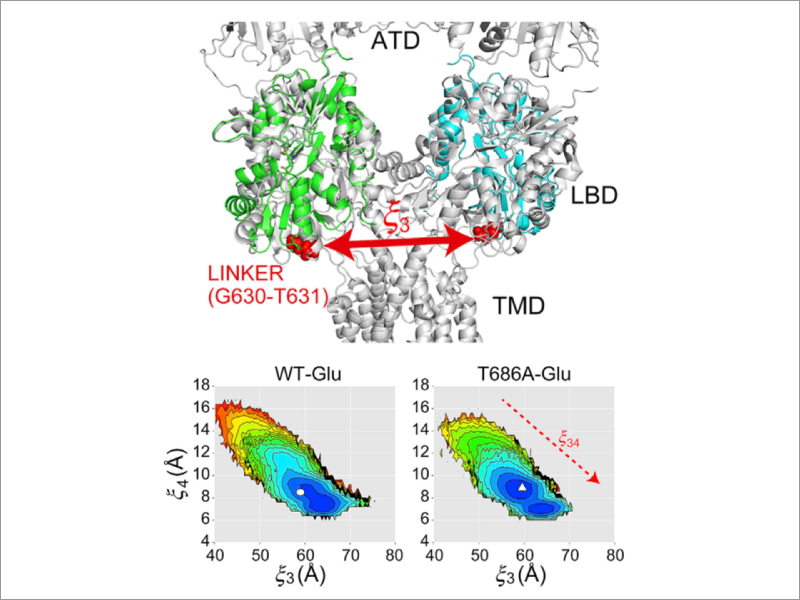
- Structural Mechanisms Underlying Activity Changes in an AMPA-type Glutamate Receptor Induced by Substitutions in Its Ligand-Binding Domain, Masayoshi Sakakura, Yumi Ohkubo, Hiraku Oshima, Suyong Re, Masahiro Ito, Yuji Sugita, Hideo Takahashi. Structure (2019) doi:https://doi.org/10.1016/j.str.2019.09.004
- Population Shift Mechanism for Partial Agonism of AMPA Receptor, Hiraku Oshima, Suyong Re, Masayoshi Sakakura, Hideo Takahashi, Yuji Sugita. Biophys. J., 116, 57-68 (2019).
Collaborators
Prof. Hideo Takahashi, Dr. Masayoshi Sakakura (Yokohama City University, Japan)