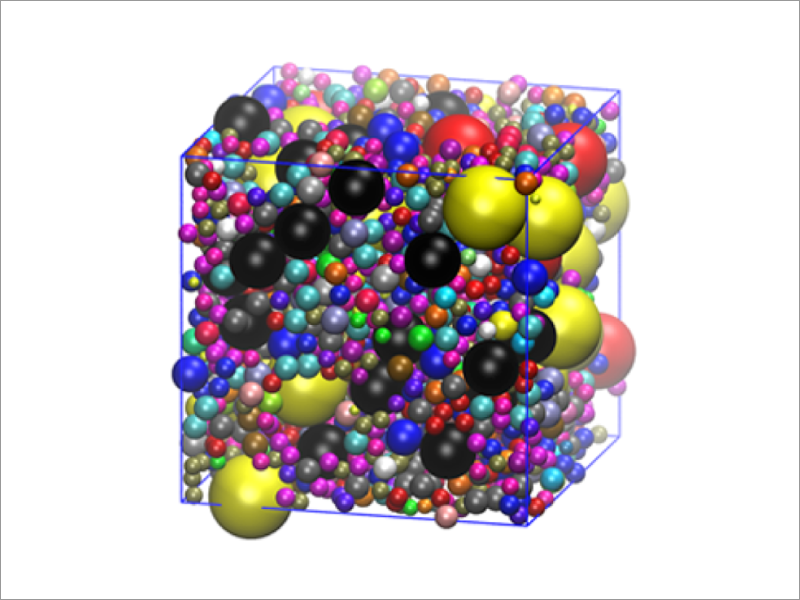
Cellular environment is significantly different from in vitro dilute environment. Inside the cell, the concentration of macromolecules, such as proteins and nucleic acids is in the range of 80–400 mg/mL. Inside the cell, proteins function by recognizing each other in a very crowded environment. It is significantly difficult, both in theory and experiment, to investigate the structure and dynamics of proteins functioning in crowded environments. The functional mechanisms of the proteins in the cell remains largely unknown at the molecular level. To simulate such crowded systems, we develop, in addition to all-atom MD simulations, multiscale models of proteins, nucleic acids and membranes, including an efficient coarse-grained model, and implement them in GENESIS. We aim to construct high-quality cytoplasmic models through multiscale simulations, and work with experimental groups such as those in the field of In-cell NMR to clarify the effects of crowded environments on biomolecule function.
References
- Thermodynamics of Macromolecular Association in Heterogeneous Crowding Environments: Theoretical and Simulation Studies with a Simplified Model. Tadashi Ando, Isseki Yu, Michael Feig and Yuji Sugita. J. Phys. Chem. B (2016) 120:11856-11865.
Collaborators
Prof. Michael Feig (Michigan State University, USA)